Ensemble learning which included multiple learners, i.e. Machine Learning algorithms, may take much longer time than expected to develop. When using a search grid for parameter optimization to train an ensemble, depending on the included algorithms, the number of variables, and the corresponding iterations based on combinations of parameter settings and with cross validation, it may take a while to produce results, assuming not running out of computing resources.
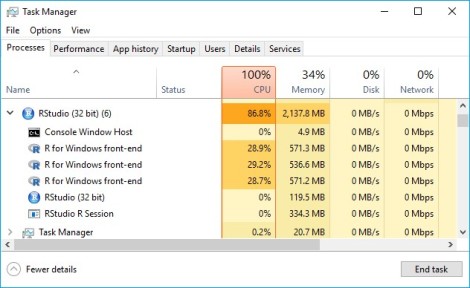
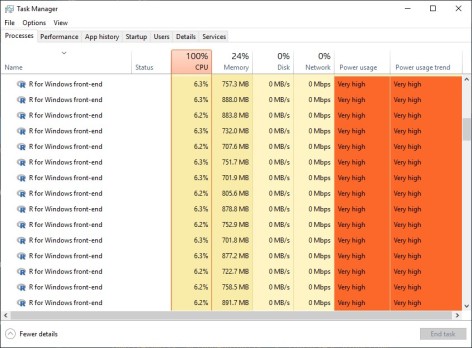
For me, parallel processing becomes essential for working on a Machine Learning project. Speaking from my experience, while developing ensemble learning with SuperLearner, in one scenario, there were total 112 learners generated from a test grid. And the wait time was just too long to maintain productivity. Later I ran parallel processing in a single host to speed up the process. I used 3 cpus of my i7-16 GB RAM laptop for small test runs and 15 vcpus of an Azure D16 Series VM with 64 RAM, as shown above, for training stable models with large amount of data. Notice that despite multiple SuperLearner sessions can run concurrently in a host with multiple cpus, within SuperLearner the process remains sequential (Ref: page 7, ‘parallel’, SuperLearner document dated Aug. 11, 2018). So using multiple cpus should and did overall reduce the elapsed time linearly (i.e. 3 cpus to cut the elapsed time to 1/3) based on my experience.
The following is one sample configuration for running parallel processing in R in Windows environment, which I employed SuperLearner for training an ensemble of ranger and xgboost. Prior to this point, I had already
- prepared and partitioned the data for training and testing (x.train, y.train, x.test, and y.test) where y is the label,
- configured the test grids (ranger.custom and xgboosst.custom) with function names resolved by SuperLearner
Upon finishing, the code also saved the run-time image as RDS object for later subsequent tasks to read in the image and eventually make predictions. Since SuperLearner does not have a built-in function to report the time for cross validation, I wrapped the cross validation part with system.time.
With the additional operation details in preparing and training an ensemble, this code is not a plug-and-play sample. If you are new to SuperLearner, I highly recommend first reviewing the package, parallel, and taking time to practice and experiment. On the other hand, if you have already had an ensemble model developed with SuperLearner, this sample code may be a template for converting existing training/model-fitting from sequential execution into a configuration for parallel processing. And stay tuned for my upcoming post, Part 2 of Predicting Hospital Readmissions with Ensemble Learning, with additional details on developing ensemble learning.
if (!require('parallel')) install.packages('parallel'); library(parallel)
# Create a cluster using most CPUs
cl <- makeCluster(detectCores()-1)
# Export all references to cluster nodes
clusterExport(cl, c( listWrappers()
,'SuperLearner' ,'CV.SuperLearner' ,'predict.SuperLearner'
,'nfold','x.train' ,'y.train' ,'x.test' ,'y.test' ,'family','nnls'
,'SL.algorithm' ,ranger.custom$names ,xgboost.custom$names
))
# Set a common seed for the cluster
clusterSetRNGStream(cl, iseed=135)
# Load libraries on workers
clusterEvalQ(cl, {
library(SuperLearner);library(caret);
library(ranger);library(xgboost)
})
# Run training session in parallel
clusterEvalQ(cl, {
ensem.nnls <- SuperLearner(Y=y.train ,X=x.train
,family=family ,method=nnls ,SL.library=SL.algorithm
);saveRDS(ensem.nnls ,'ensem.nnls')
})
# Do cross validation in parallel
system.time({
ensem.nnls.cv <- CV.SuperLearner(Y=y.test ,X=x.test
,cvControl=list(V=nfold) ,parallel=cl
,family=family ,method=nnls ,SL.library=SL.algorithm
);saveRDS(ensem.nnls.cv ,'ensem.nnls.cv')
})
stopCluster(cl)
